
Tsvetanova NG, Klass DM, Salzman J et al (2010) Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. Scherrer T, Mittal N, Janga SC et al (2010) A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. Lunde BM, Moore C, Varani G (2007) RNA-binding proteins: modular design for efficient function. Greenberg JR (1979) Ultraviolet light-induced crosslinking of mRNA to proteins. Gerstberger S, Hafner M, Tuschl T (2014) A census of human RNA-binding proteins. Kaper JM (1969) Nucleic acid-protein interactions in turnip yellow mosaic virus. Moreover, we provide notes on experimental design and a troubleshooting guideline for common problems that can occur during iCLIP library preparation. In this chapter, we outline the iCLIP protocol and list possible controls that allow a targeted and cost-minimizing optimization of the protocol for an RBP-of-interest. Here, we summarize individual-nucleotide resolution UV cross-linking and immunoprecipitation (iCLIP), a powerful technique that provides genome-wide information on RNA–RBP interactions at nucleotide resolution. Furthermore, the development of high-throughput sequencing has broadened the capability of these methods. Several techniques have been developed to capture the properties of RNA–RBP interactions. Gaining deeper insights into the biology of RNA–RBP interactions will lead to a better understanding of regulatory processes and disease development. Consistently, mutations in RBPs result in defects in developmental processes, diseases, and cancer. The regulatory mechanisms are controlled by RNA-binding proteins (RBPs), which form complexes with RNA and regulate RNA processing, stability, and localization, among others.
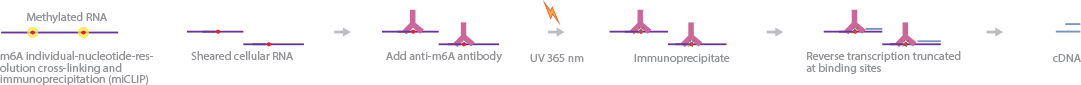
The importance of posttranscriptional regulation in cellular metabolism has recently gone beyond what was previously appreciated.


 0 kommentar(er)
0 kommentar(er)
